The current tree of life has three main branches: archaea, bacteria, and eukaryote. This three-domain system was introduced back in 1977. As proposed by researchers (Woese et al., University of Illinois), the basis of the classification relies on differences in 16S rRNA genes. Guess which form of (arguably) life that does not have rRNA? Viruses. That’s why it is not on the tree of life and that’s why for some people viruses are not living things. Viruses are just particles that can interact, sometimes in nasty ways, with other living things.
But what if there is actually a fourth domain of cellular life? Researchers are investigating the possibility of a fourth domain, however the evidence we have so far are not compelling to establish a new domain of life. Interestingly, works are being done to embrace such possibility because virologists begin to take a peek into a new world of microbes: the giant viruses, also known as giruses. After reading a paper by researchers from the NCBI (Schulz et al., DOI:10.1126/science.aal4657), I was entranced by it. There was one sentence that captured my attention:
“It has been proposed that giant viruses constitute remnants of ancient cellular life or are derived from an enigmatic fourth domain of life”
The researchers set up an experiment to test the fourth domain hypothesis. They did so by performing a low-complexity metagenomic analysis from a wastewater treatment plant in Klosterneuburg, Austria. Metagenomics means that a direct genetic analysis of genomes contained within an environmental sample are directly performed. In other words, take a sample, purify the DNA, then sequence them all. It eliminates the step for growing samples in culture because some bacteria and viruses just can’t be grown in lab.
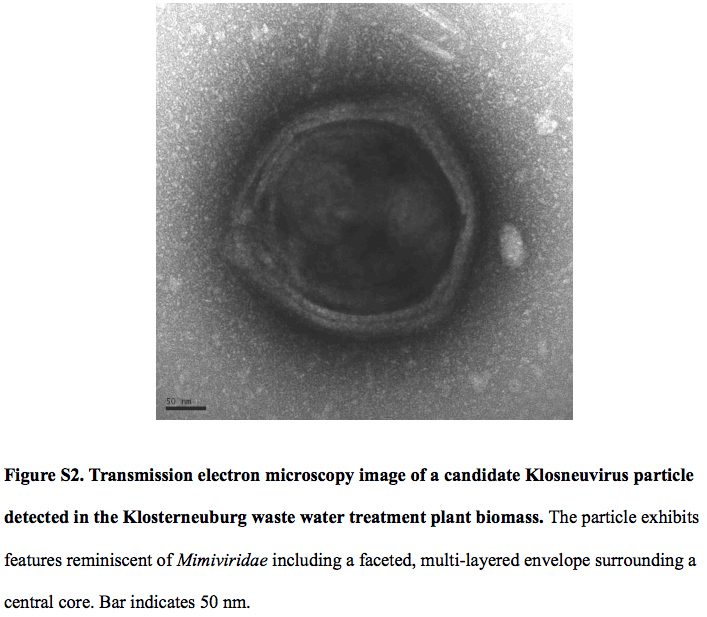
Their data shows a 1.57-Mb genome of a putative virus named Klosneuvirus. Klosneuvirus is the subject of interest here because it can code for an expanded translation machinery that includes aminoacyl tRNA synthetases (aaRSs) with specificities for all 20 amino acids. What does an aaRS do? It attaches the appropriate amino acid onto its tRNA, allowing the translation process to occur. Why this is a big deal? Because most viruses do not have their own factory to carry out translation process because they rely on (read: steal) the host’s protein-making machinery. In fact, protein-making machinery is too bulky for a small virus to carry it together in its genome.
The first girus I learned was Mimivirus, thanks to the instructor for my virology course (fall 2015). This paper mentions that even Mimivirus does not have such a sophisticated translation machinery. It also mentions that Klosneuvirus and Mimivirus share about 200 gene families. Klosneuvirus has 355 genes, with 12 of them being shared with eukaryotes but not with other viruses. This means that Klosneuvirus probably acquired those genes through its dynamic interaction with its host.
So what about the fourth domain hypothesis now? Has Klosneuvirus demonstrated the hypothesis being compatible in this regard? Well, the answer seems to be a no. A comprehensive phylogenomic analysis was performed and they found out that the aaRSs and various translation factors are related with diverse eukaryotes, mainly protists. It means that probably in the past the Klosneuvirus acquired genes from the protists it infected. If the fourth kingdom hypothesis is true, we would expect that the aaRSs and translation factors encoded by Klosneuvirus to be distinct from that of protists.
It would be interesting if the fourth domain hypothesis is scientifically proven as true. It means that we need to learn a lot of things about life itself.
Sources and more reading materials:
- Phylogenetic structure of the prokaryotic domain: The primary kingdoms. DOI:10.1073/pnas.74.11.5088
- Let There Be Life | TheScientist
- Metagenomics - a guide from sampling to data analysis, DOI:10.1186/2042-5783-2-3
- Nucleocytoplasmic large DNA viruses (NCLDV), order Megavirales. Wikipedia entry here
- Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life, DOI:10.1016/j.virol.2014.06.032