Alphabet soup is remotely fun. Here is a mini-note about the interaction between spacer DNA sequence, the transcribed CRISPR crRNA, the target DNA, and the protospacer adjacent motif sequence. We are going to assume that the context of our situation for all these terms is not in eukaryotic gene editing but as for bacterial adaptive immune response.
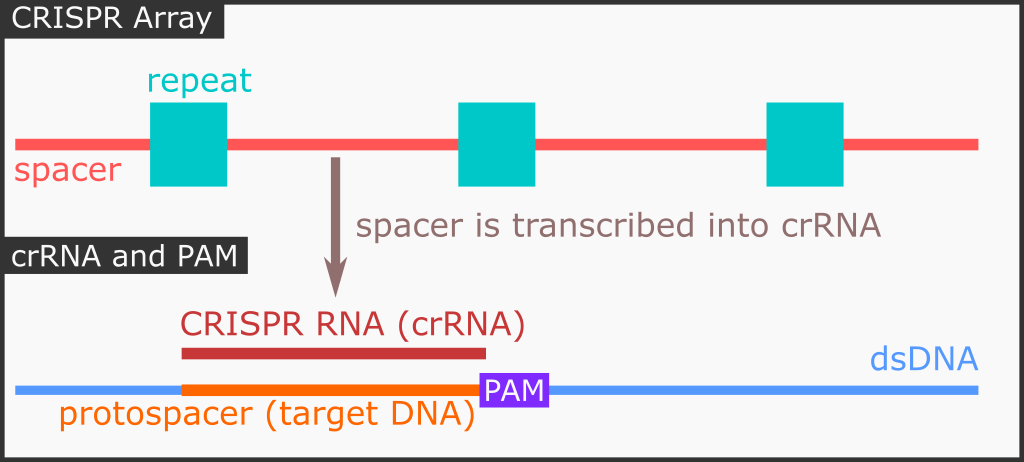
Previously incorporated invading phage DNA sequence is stored as spacer sequence, flanked by repeating sequence (we just call it the “repeat” sequence). Together, they are known as the repeat-spacer array. The next step is that we need to transcribe the spacer into something that Cas9 can utilize: CRISPR RNA (crRNA). To summarize, from spacer sequence we now have the crRNA.
Now that our Cas9 protein can utilize the crRNA. However, crRNA needs to form a complex with a trans-activating CRISPR RNA (tracrRNA) for the crRNA to be “active”. We are not going to discuss that in depth here.
The crRNA should be complement with the target DNA. The short, 20-nt, target DNA sequence must be adjacent with a protospacer adjacent motif (PAM) sequence. I was having a hard time why it is called a PAM sequence, especially the word protospacer. It turns out here is the story.
When an invading phage DNA sequence is intercepted by the CRISPR/Cas foreign DNA acquisition machinery, Cas (not necessary the Cas9, can be other Cas proteins) will locate PAM sequence on the foreign DNA sequence and then will acquire a new sequence to be incorporated into the repeat-spacer array. As you might have guessed, the sequence to be acquired here is the spacer. Prior to being acquired, it is called protospacer. It makes sense because proto means original.
One more thing. The crRNA does not interact with the PAM sequence, the Cas9 protein does. It is like the Cas9 protein (and any other related Cas proteins) has an innate capability to recognize certain specific sequence prior to cleaving.
Virginijus Siksnys and his team did a study on this. It is pretty interesting.