I have mastered the technique to make tabloidy headlines.
No seriously.
Yesterday I saw a news where researchers at the MIT Broad Institute led by Dr. Feng Zhang published a work on a nucleic acid detection system called SHERLOCK, which stands for Specific High-Sensitivity Receptor Unlocking (doi:10.1126/science.aam9321). To be honest, its long name makes no sense to me but because its short name is cool, so I will leave it at that.
What SHERLOCK does? Let’s get to the problem statement first. Consider a case where we need to detect and distinguish between zika (ZIKV) and dengue (DENV) infection. These two viruses pose challenge when it comes to detection because they are quite similar, genome-wise and antibody-wise. Sometimes researchers can’t tell between those two because antibody assay against DENV could react with ZIKV. There is one interesting paper on ZIKV and DENV where previous infection with DENV could drive an antibody-dependent enhancement of ZIKV (doi:10.1038/ni.3515), a.k.a the infamous problem known as the original antigenic sin.
SHERLOCK aims to solve that. To prove SHERLOCK’s detection specificity, the researchers produced lentiviral constructs with nucleic acid fragments from ZIKV and DENV. With SHERLOCK, these two fragments can be distinguished down to 2 attomolar (aM) concentration. That’s really something. We need this specificity in the field because that eliminates the need for polymerase chain reaction (PCR), thus simplifying the detection protocol.
What makes SHERLOCK as a potential point-of-care (POC) device? The paper mentions in its abstract that this CRISPR-based diagnostic (a.k.a CRISPR-Dx), the SHERLOCK, can be lyophilized (a.k.a freeze-dried) for a long-term storage in absence of a cold chain system, which is a big deal in poor countries where there is no electricity to run freezers. On top of that, all SHERLOCK components are combined in a single reaction, making it a great platform for POC. How so? SHERLOCK can be run in the field by using the paper-spotting protocol and it could detect 20 femtomolar (fM) of non-amplified DNA. Again, no PCR needed.
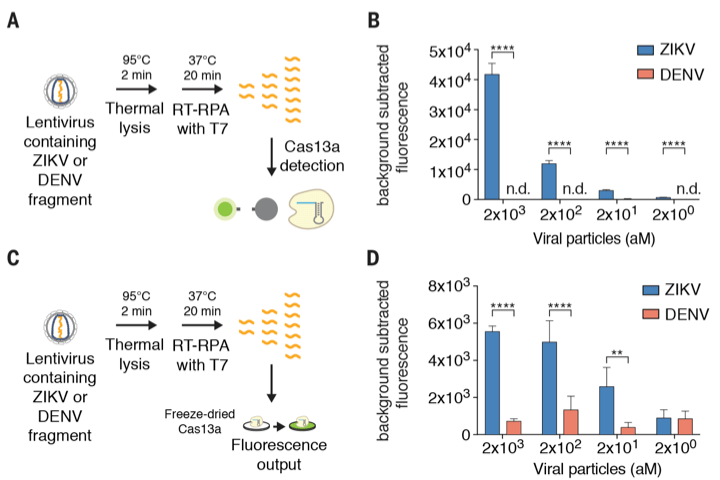
We now know that SHERLOCK has a great potential, what is it actually? Previously described as C2c2 Cas protein, it is now known as the Cas13a. Unlike typical Cas proteins in previous publications, Cas13a uses crRNA to target RNA instead of DNA. In nature, the presence of Cas13a provides bacteria with the capability fight against RNA phages.
At first I was hung up when reading this paper. Look at this figure 1-A.
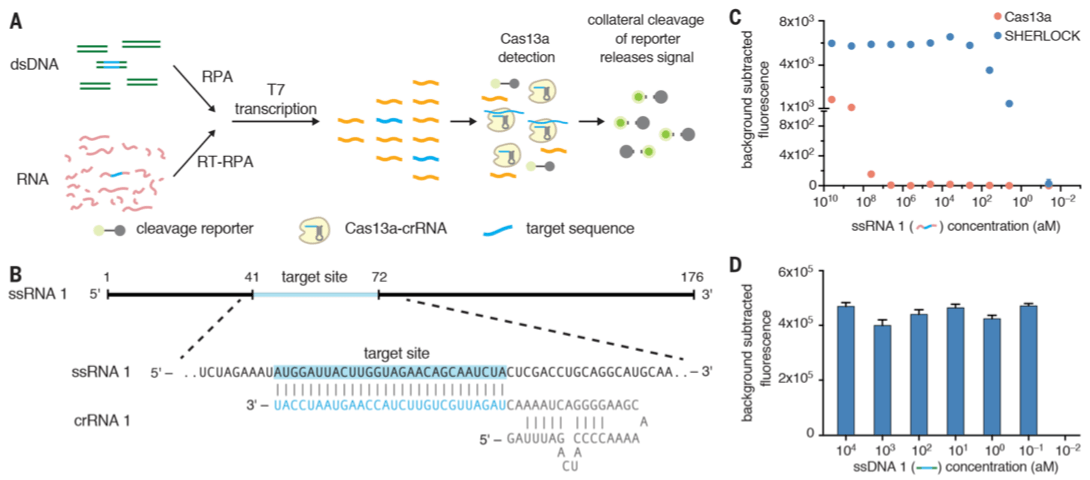
I had this question in my mind. If this is done out in the field, how do we know the reaction is successful? They have a reporter (a quenched fluorescence molecule on a RNA fragment) there, so how does this reporter fit in a real-world situation? Turns out, Cas13a has an activity termed as collateral cleavage. If the crRNA that guides the Cas13a finds a match (in this case it would be a viral genome), the Cas13a will first cut the target genome. However, it won’t stop there. In a paper published by Zhang’s lab back in June 2016 (when it was still called as C2c2, doi:10.1126/science.aaf5573), Cas13a is described to have a collateral cleavage activity where it continues to cut RNA fragments non-specifically. This is where the RNA reporter sequence plays a role.
For example, if the crRNA:Cas13a complex recognizes a target viral genome, Cas13a first cleaves the target and then it stays active, cutting RNA fragments that includes the RNA reporter sequence. The design of the RNA reporter sequence merits the name SHERLOCK. I still do not understand the “receptor” part anyways, but the “unlocking” part makes sense. It is because a fluorescence molecule is “locked” (quenched) when there is a quencher molecule nearby, a concept known as Förster resonance energy transfer (FRET). When both fluorescence and quencher molecules are separated, the quencher molecule can’t no longer lock the fluorophore, thus it emits light. Neat!
Couple things I learned from this paper:
- digital-droplet PCR (ddPCR), is improved version of PCR with single-molecule sensitivity. How so? By partitioning samples into droplets, ddPCR makes it easier for quantification. Too hard to understand? Watch this video.
- Recombinase polymerase amplification (RPA) is the isothermal alternative to the PCR. It requires a reverse transcriptase (RT). The great thing is that RPA could be used to detect RNA and DNA without cDNA intermediate. Since it is an isothermal procedure, no thermal cycler is needed and it works best at 37-42 degree Celsius.
By the way, take a look at the SHERLOCK paper and C2c2 paper and pay a closer attention to the first two authors.